Algorithms and Applications of Novel Capsule Networks
- Rodney LaLonde

- Jul 12, 2020
- 23 min read
Hello Everyone! This is going to be a much longer blog post than usual, but I will try to summarize my PhD Defense on the Algorithms and Applications of Novel Capsule Networks in as few words as possible. Everything in this post is covered in the video of my defense which you can watch here.
Chapter 1: Background & Motivation
"Convolutional Neural Networks are doomed." – Geoffrey Hinton
This is a famous quote from Geoffrey Hinton, Turing award winner and often called the godfather of deep learning, taken from his 2013 lecture series on computer vision as inverse graphics. Let’s see why Hinton proclaimed this…
Major Shortcomings of CNNs
Convolutional neural network (CNN) features are scalar and additive:

Fig. Credit: https://cs231n.github.io/convolutional-networks/
This leads to two major limitations.
The modeling of internal relationships amongst neurons becomes ambiguous when we can only sum presence estimations over receptive fields, a problem which is made worse by non-overlapping subsampling operations such as the Max Pooling operation.
CNNs lack the ability to extrapolate to new viewpoints. Since features are represented as scalar values, the only information they can contain is the probability of whether or not a feature is present at a given location.
Hinton famously said about the first problem,
"The pooling operation used in convolutional neural networks is a big mistake and the fact that it works so well is a disaster." – Geoffrey Hinton
We can demonstrate this shortcoming with a quick example.

Given this scrambled face as input to a CNN for classification, early works such as Zeiler and Fergus in 2013 show CNNs learn high-value localized activations on mid-level feature maps. Passing those features into the higher-level layers, we would obtain a feature map which gets classified as a face, despite the input being all jumbled up.
The second, and perhaps more important limitation, is CNNs lack the ability to extrapolate to new viewpoints. Since features are represented as scalar values, the only information they can contain is the probability of whether or not a feature is present at a given location. This means we must either replicate our feature detectors across all possible poses of all possible inputs at every single layer (something we can easily see is computationally infeasible), or we train our CNNs in such a way as to become invariant to all possible pose changes of each feature. This lattermost method is typically what is done today through the use of data augmentation, and while it provides some flexibility, it’s largely ineffective. Here is a simple demonstration of these issues played out in practice.

Fig credit: https://hackernoon.com/capsule-networks-are-shaking-up-ai-heres-how-to-use-them-c233a0971952.
At left, the network is now even more confident that our disarranged face belongs to a person, while at right, simply rotating the image upside down causes it to be misclassified as coal. In fact, this issue is so extensive, that a 2019 study presented at CVPR found CNNs fail to recognize 97% of the pose space of objects.

Figure credit: Alcorn, Michael A., et al. "Strike (with) a pose: Neural networks are easily fooled by strange poses of familiar objects." CVPR 2019.
Capsule Networks to the Rescue
Now, capsule networks are an attractive potential solution to these shortcomings in the CNN literature.

A capsule network is comprised of capsules, which are defined as a group of neurons representing a feature’s presence, and, more importantly, the attributes of that feature, often called the feature’s instantiation parameters, and these can be anything from pose to deformations, to velocity in video to albedo, hue, texture, and many other things.
In practice, this only requires two simple changes from convolutional neural networks:
Features are now represented by vectors or matrices rather than scalars and this allows us to store important information about the features being learned.
Information is routed from one layer to the next via a dynamic routing algorithm which weights both the presence and internal relationships of the lower-level features with respect to each higher-level combination.
We can illustrate how this solves the previously discussed problems with a quick example.

If we take our jumbled face image and input it to a capsule network, we would obtain vectors whose dimensions contain the instantiation parameters necessary to construct the features they represent.

After passing these vectors, now represented here by their reconstructions, to the transformation matrix for the face parent capsule, this produces a set of prediction vectors. Here, each child capsule is making a loud vote that a face should be present, but the pose of that face is in strong disagreement amongst the different children, shown by the non-overlapping faces. After sending these predictions through the dynamic routing algorithm, this disagreement causes no activation of the face capsule.

If instead we input a proper face image, now the children’s votes all agree on the pose of the face that’s present. And when combining these predictions in the dynamic routing algorithm, we would observe an activation of the face capsule with the face’s instantiation parameters captured as well.
Practical Benefits of Capsule Networks
With this understanding of capsule networks, let’s examine some practical benefits of adopting this framework.

Since we’re storing richer information about features, and because of the combination of transformation matrices and dynamic routing, we can now generalize our features across pose variations, even to those which are unseen during training.
Essentially what we’re doing is imposing a coordinate frame on our features and then the multiplication of those features with a transformation matrix equates to a viewpoint change.
This means we no longer need to massively replicate feature detectors across viewpoints to generalize to new poses of objects, which we demonstrate in one of our works.
Therefore, it follows, that we should be able to achieve the same levels of predictive performance of very deep CNNs, while only using some small fraction of their parameters due to these much stronger internal representations being learned; in fact, we find this to be true in all of our published capsule works.
And thus, in culmination, due to having fewer parameters and being able to generalize across viewpoints, we should find that capsule networks will require far less data to train while converging significantly faster.
In “Capsule networks against medical imaging data challenges”, the authors show that capsule networks require less data to train as well as handle class imbalances better than CNNs, and we observe faster convergence in all of our studies.
[1] Jiménez-Sánchez, Amelia, Shadi Albarqouni, and Diana Mateus. "Capsule networks against medical imaging data challenges." Intravascular Imaging and Computer Assisted Stenting and Large-Scale Annotation of Biomedical Data and Expert Label Synthesis. Springer, Cham, 2018. 150-160.
Research Questions
How effective are capsule networks in real-world applications? *
What algorithmic advances are needed for these new applications?
* Specific focus is given to biomedical image applications for their significance in potentially life-saving technologies.
Chapter 2: Capsules for Image Segmentation
First-ever capsule-based segmentation network. Related Publications and Patent:
Rodney LaLonde and Ulas Bagci. "Capsules for Object Segmentation." Medical Imaging with Deep Learning (MIDL 2018). (Oral; CIFAR Travel Award for Outstanding Papers; ~110 citations).
Rodney LaLonde, Ziyue Xu, Sanjay Jain, and Ulas Bagci. "Capsules for Biomedical Image Segmentation." Elsevier, Medical Image Analysis (Under Revision). 2020.
Rodney LaLonde and Ulas Bagci. "Capsules for Image Analysis." U.S. Patent Application 16/431,387; filed December 5, 2019.
Sumit Laha, Rodney LaLonde, Austin E. Carmack, Hassan Foroosh, John C. Olson, Saad Shaikh, and Ulas Bagci. "Analysis of Video Retinal Angiography with Deep learning and Eulerian Magnification." Springer, Frontiers in Computer Science. 2020.
Motivation and Challenges
The equivariant properties of capsule networks lend themselves nicely to segmentation, where precise spatial localization is very important. However, the initial CapsNet published by Sabour, Frosst, and Hinton in 2017 was extremely computationally expensive. Given just a small 6 by 6 pixel grid of 32 8-dimensional capsules being routed to 10 16-dimensional capsules for classification, there are nearly 1.5 million parameters. With the task of segmentation requiring far larger input and output sizes (e.g. 512 × 512 pixels), the number of parameters quickly grows out of control, making it impossible to fit such models into memory.
512×512×32×8×512×512×10×16 = 2,814,749,767,106,560 parameters.
To solve the memory burden, we introduce two important contributions. First, we propose a locally-constrained dynamic routing algorithm to route information to each parent capsule only from a small local neighborhood of child capsules centered on that parent’s spatial location. Second, we propose to share transformation matrices across both spatial locations and child capsule types. These two contributions combined, reduces the number of parameters in every capsule layer by a significant factor, reducing those needed 2.8 quadrillion parameters to only 324 thousand.
Reduces parameters by 𝑪×𝑯×𝑾×𝑯⁄𝒌 × 𝑾⁄𝒌. 2.8 quadrillion parameters cut down to 324,000 (with 𝑘=5).
Locally-Constrained Dynamic Routing and Transformation Matrix Sharing
In Sabour et al. 2017, Prediction vectors, 𝒖̂, are created for every child capsule, i, to every parent capsule, j.

In our proposed locally-constrained dynamic routing with transformation matrix sharing, prediction vectors, 𝒖̂, are only created from a 𝑘×𝑘 kernel of child capsules, 𝑖, within each child capsule type, centered at the (𝑥,𝑦) position of each parent capsule, 𝑗. Transformation matrices are shared across the spatial grid and child capsule types to create predictions for each parent capsule.

Reconstruction Regularization
Another component of the original CapsNet, was a learned an inverse mapping from capsule vectors back to inputs, as a form of regularization. To construct a similar regularization technique for segmentation, we modify the algorithm to reconstruct all input pixels belonging to the positive input class, using the ground-truth at training and all capsules whose vector lengths are above a given threshold at testing. This learned inverse mapping forces the capsule vectors to learn the instantiation parameters of objects, while the main loss enforces learning discriminative classification features. This technique shares parallels to works such as VEEGAN which learned an inverse mapping to overcome the mode collapse issue in GANs.

Baseline Segmentation Capsule Network
Putting these 3 novelties together, we can create our first capsule network for object segmentation, which serves as a baseline for us. You’ll notice the structure is kept virtually identical to CapsNet, with the dynamic routing swapped out for our locally-constrained routing with transformation matrix sharing, and the new reconstruction method added. The images shown as input and output are from the task of retinal vessel segmentation from the fluorescein angiogram videos.

Loss of Global Information & Its Recovery
While we saw good performance on the retinal vessel segmentation task, locally-constraining the dynamic routing introduces a major limitation. Segmentation as a task is really the joining of two separate tasks solved in unison: recognition and delineation. This is why most successful segmentation frameworks utilize encoder-decoder networks, to obtain global information for recognition and local information for delineation. When we constrained the dynamic routing, we lost the ability of capturing global information at each layer. To solve this, we introduce a novel encoder-decoder style capsule architecture, by creating “deconvolutional” capsules. These deconvolutional capsules operate in the same manner as our convolutional capsules, except their prediction vectors are formed using a transposed convolution operation. Now, with these five novelties combined, namely the locally-constrained dynamic routing, transformation matrix sharing, reconstruction regularization for segmentation, deconvolutional capsules, and our deep encoder-decoder capsule architecture, we produce the first ever capsule-based segmentation network in the literature, called SegCaps.

Datasets and Experiments: Pathological Lung Segmentation
For this work, we performed the largest pathological lung segmentation study in the literature, combining five large scale datasets from both clinical and pre-clinical subjects. With the goal of creating a general framework which performs consistently across many different lung diseases and even different anatomies, we conducted experiments on the following datasets: LIDC, containing lung cancer screening patients; LTRC, containing interstitial lung disease, specifically fibrotic, as well as COPD patients; UHG, containing 13 different forms of interstitial lung diseases; JHU’s TBS, which looks at the effect of smoking inhalation on the development and progression of tuberculosis in mice subjects; and JHU’s TB, which looks at the effect of two different experimental treatments on the development and progression of TB in mice subjects.

It’s worth noting that preclinical image analysis is a particularly challenging area, with extremely limited training data due to a lack of expert interest in providing annotations (I myself actually spent over 2 months annotating data to complement the annotations provided by radiologists). Combining that with drastically different anatomy and extremely high levels of noise, it quickly becomes a significantly difficult task. No works for preclinical segmentation exist, beyond Dr. Ulas Bagci’s 2015 TMI paper, a semi-automated non-deep machine learning method.
For all experiments, we compared five methods: U-Net, the gold standard in biomedical image segmentation for the last several years; Tiramisu, a dense encoder-decoder extension to U-Net; P-HNN, the state-of-the-art method in pathological lung segmentation; our baseline SegCaps model; and the proposed SegCaps. Shown in the right-hand column is a comparison of the number of parameters used in each of these networks, where our proposed SegCaps contains less than 5% of the parameters, at only 1.4 million, of the typical U-Net.

Pathological Lung Segmentation Results
These are the results of our experiments. We computed the dice coefficient, which captures the global-level overlap of segmentations with the ground truth, and the Hausdorff distance, which captures the local-level accuracy of the segmentation boundaries. In clinical and preclinical subjects, SegCaps is consistently outperforming all other methods despite using only a small fraction of the size of these bigger networks. Since the lungs are such a large portion of the input space, the true comparison of accuracy between methods is best captured by the Hausdorff distance. Where the Dice scores are fairly close for all methods, the Hausdorff distance is typically only closely clustered for the CNN-based methods, with SegCaps significantly outperforming them in most datasets.





Retinal Vessel Segmentation Results
Next we will look at some of our work done on retinal vessel segmentation. These qualitative results demonstrate the superiority of our baseline capsule network over U-Net, particularly on thin vessels, seen here, or crowded vessels, seen here. Since this task doesn’t require global information we chose not to use our encoder-decoder structure. In the results, areas of cyan are being under-segmented by the method, magenta areas are being over-segmented, and white areas are correct segmentations. U-Net struggles with issues of under-segmentation on the thin vessel structures, and over-segmentation in areas of crowded vessels. Overall our capsule-based segmentation network achieved consistently better results than U-Net.

Can Capsules Really Generalize Better to Unseen Poses than CNNs
Our last set of experiments examined the ability of SegCaps to generalize to unseen poses, an understudied but purported benefit inherent to capsule networks. We overfit U-Net and SegCaps to 100% training accuracy on a single image, then rotated or reflected that image and fed it to the networks for prediction. U-Net struggled to provide good segmentations, while SegCaps saw only a minor drop in performance. Shown in the chart at right, U-Net’s performance dropped nearly 15% while SegCaps only dropped around 4% on average. To guarantee a fair comparison and ensure the significantly more parameters in U-Net had a chance to properly fit, we trained both networks for 10 times the number of epochs past convergence. U-Net still struggled to handle these changes in viewpoint, showing nearly the same degree of performance drop, while SegCaps handled them again with relative ease, now showing almost no drop in performance.

Conclusions and Discussion
The proposed framework is the first use of the recently introduced capsule network architecture and expands it in several significant ways. First, we modify the original dynamic routing algorithm to act locally when routing children capsules to parent capsules and to share transformation matrices across capsules within the same capsule type. These changes dramatically reduce the memory and parameter burden of the original capsule implementation and allows for operating on large image sizes, whereas previous capsule networks were restricted to very small inputs. To compensate for the loss of global information, we introduce the concept of “deconvolutional capsules” and a deep convolutional-deconvolutional capsule architecture for pixel level predictions of object labels. Finally, we extend the masked reconstruction of the target class as a regularization strategy for the segmentation problem.
Experimentally, SegCaps produces improved accuracy for lung segmentation on five datasets from clinical and pre-clinical subjects, in terms of Dice coefficient and Hausdorff distance, when compared with state-of-the-art networks U-Net (Ronneberger et al., 2015), Tiramisu (Jegou et al., 2017), and P-HNN (Harrison et al., 2017). More importantly, the proposed SegCaps architecture provides strong evidence that the capsule-based framework can more efficiently utilize network parameters, achieving higher predictive performance while using 95.4% fewer parameters than U-Net, 90.5% fewer than P-HNN, and 85.1% fewer than Tiramisu. To the best of our knowledge, this work represents the largest study in pathological lung segmentation, and the only showing results on pre-clinical subjects utilizing state-of-the-art deep learning methods.
To demonstrate the extended scope and potential impact of our study, we have performed two additional sets of experiments in object segmentation: 1. Segmenting retinal vessels, containing extremely thin tree-like structures, from retinal angiography video. 2. Testing the affine equivariant properties of SegCaps on natural images from PASCAL VOC. The results of these experiments, as well as the main body of our study, demonstrate the effectiveness of the proposed capsule-based segmentation framework. This study provides helpful insights into future capsule-based works and provides lung-field segmentation analysis on pre-clinical subjects for the first time in the literature
SegCaps introduced many important novelties to create the first-ever capsule segmentation network. However, there is more to medical imaging applications than just segmentation. The obvious next step, is to create a capsule-based network for computer-aided diagnosis, where an end-to-end detection and diagnosis system is the ultimate goal. Although capsule networks do exist for classification tasks, there are still some major limitations to be overcome for their use in real-world applications.
Chapter 3: Capsule-Based Medical Diagnosis
Creating a capsule based network for medical image diagnosis. Related Publications and Patent:
Rodney LaLonde, Pujan Kandel, Concetto Spampinato, Michael B Wallace, and Ulas Bagci. "Diagnosing Colorectal Polyps in the Wild with Capsule Networks." IEEE International Symposium on Biomedical Imaging (ISBI 2020).
Pujan Kandel, Rodney LaLonde, Victor Ciofoaia, Michael B Wallace, and Ulas Bagci. "Colorectal Polyp Diagnosis with Contemporary Artificial Intelligence." Elsevier, Gastrointestinal Endoscopy, Volume 89, Issue 6. 2019.
Rodney LaLonde and Ulas Bagci. "Capsules for Image Analysis." U.S. Patent Application 16/431,387; filed December 5, 2019.
Background, Motivation, and Challenges
While the memory saving techniques introduced in SegCaps allow us to perform segmentation on high-dimensional inputs, classification at these sizes still remains an issue. Existing capsule-based frameworks for medical diagnosis utilize CapsNet’s original fully-connected capsules, forcing these methods to rely on inefficient sliding-window style approaches to handle the larger input sizes in medical imaging. To overcome this, we introduce a capsule-average pooling algorithm, to create a more efficient classification network. Our chosen application is colorectal cancer diagnosis on poorly-localized, “in-the-wild”, polyps.
First, some quick details about this chosen application.

Colorectal cancer is the third most common cancer diagnosed in men and women, and is the second highest cause of cancer deaths in the United States. Typically, colorectal cancer begins as polyps which can grow into invasive carcinomas. Each year more than 140 thousand people are diagnosed, and there has been a recent increase in the diagnosis of people under the age of 50. And, most importantly, catching it early significantly increases the patients chance of survival, with only about a 10% survival rate if not found until a later stage.
Colorectal Polyps "In-The-Wild"
Previous studies on colorectal polyps have focused on limited, well-curated datasets, such as the one shown in the video below.
While these studies show promising results, polyps are not always perfectly localized with thousands of frames of video available for every single polyp. In this study, we choose to focus on more diverse and challenging data, illustrated by the two examples shown below, with the red arrows highlighting the polyps to be diagnosed.

Looking at this in a bit more detail...

Previous datasets contain a limited numbers of polyps, typically with not much variation in they way they are captured, and a large amount of data for each polyp, with usually over 1600 frames of close-up video all providing complimentary information from multiple viewing angles, modalities, focus modes, and lighting conditions, to help create an optimal diagnosis. Our in the wild dataset, contains far more, typically non-localized polyps, with just a single image per imaging mode, and often only a single imaging mode provided, to make our prediction for a diagnosis. Combining these challenges with the large scale, skew, and illumination changes, shown in the examples on the previous slide, and this becomes an incredibly difficult task.
Which brings us to our hypothesis for this chapter...
"Given the preliminary evidence that capsule networks can better generalize to unseen poses and requires less data for training, we hypothesize that a capsule-based diagnosis network should be able to better handle the relatively limited training data and high intra-class variation present in this colorectal polyp dataset."
Capsule-Average Pooling (CAP)
To perform diagnosis on our high-dimensional imaging data, without resorting to inefficient sliding windows, we implement a capsule-average pooling layer to create a more efficient classification network, in similar manner to how global average pooling layers work in CNNs. We reduce the dimensionality of our features by computing the average capsule activations and poses, across spatial dimensions and within capsule types. Then, we restructure these into a single set of class-capsule prediction vectors. This works by computing the vector mean across the height and width of the capsule grid, preserving the length of capsule vectors within each capsule type. Finally, we perform diagnosis by taking the magnitude of our restructured class prediction capsules.
Diagnosis Capsule (D-Caps)

We call our capsule-based diagnosis architecture, D-Caps. First, we input a high-dimensional image of a colorectal polyp, to a relatively deep capsule network made entirely of capsule layers. At the end of the network, we apply our proposed capsule-average pooling layer, which forms our class prediction capsules. Following this, the magnitude of each capsule is computed to predict the output diagnosis, while the highest scoring class capsule is sent to the reconstruction subnetwork for regularization.
Dataset and Experiments
To test our method, experiments were conducted on the Mayo Polyp dataset collected at the Mayo Clinic, Jacksonville by Dr. Michael B. Wallace his colleagues. It contains 963 polyps collected from standard and dual-focus colonoscopes with both white light and narrow-band imaging settings. We conducted three sets of experiments with D-Caps and Inceptionv3 (Iv3):
Hyperplastic versus Adenoma, which is the most common experiment conducted in the literature;
Hyperplastic vs Adenoma and Serrated, which is clinically meaningful as hyperplastic polyps are benign can be safely left in situ while both adenoma and serrated polyps can become cancerous and must be resected;
Hyperplastic versus Serrated, where serrated polyps are typically left out of most studies since they can appear visually similar to hyperplastic polyps, causing many automated algorithms to have difficulty in distinguishing them.



The results of our experiments show that D-Caps is able to significantly outperform the previous state-of-the-art method based on inceptionv3, improving performance by 17, 27, and 43% relative accuracy increases on these three tasks of increasing difficulty at the polyp level.
We also conducted ablation studies on the components of our proposed method, and found that:
In our routing iteration ablation experiment, we obtained 50.61%, 65.53%, 45.97%, and 50.86% accuracy at the polyp level for 2, 3, 4, and 5 routing iterations respectively.
Removing the reconstruction sub-network obtained 56%, 50%, and 55% accuracy at the for experiments 1 – 3 respectively, an average 8% decrease.
Localization is of critical importance for obtaining an optimal diagnosis. On an ideal subset of physician chosen images, we obtained an accuracy of 82% for hyperplastic vs adenoma. For this, we asked our collaborating physician to select a subset of 100 more “ideal” cases, where polyps are better localized in the frame by the colonoscopist; although still only given a single image, we obtained significantly improved results, getting closer to clinically acceptable levels.
Conclusions and Discussion
These experiments show the dynamic routing and reconstruction both contribute to the overall performance of our model, while the latter experiment provides strong evidence that with further improvements in both capsule network algorithms and screening technology, AI-driven approaches can prove viable for raising optical biopsy techniques to clinical practice standards. Our work provides an important baseline for future studies on the extremely challenging Mayo Polyp dataset, and contributes further evidence that given limited data with high intra-class variation, capsule networks can significantly outperform deep CNNs.
In this chapter, and in the medical imaging domain in general, we often have limited training data, with potentially a large degree of variation within, and not across classes. For example, think of the intra-class and inter-class variation for classifying cats vs airplanes, then the intra- and inter-class variation of malignant vs benign polyps. D-Caps represents an important step towards a more clinically-viable computer aided-diagnosis system, showing it can handle these types of real-world challenges better than a state-of-the-art CNN. However, there is still a more significant barrier to the adoption of computer-aided diagnosis systems into routine clinical workflows.
Chapter 4: Encoding Capsules for Explainable Predictions
Creating an explainable multi-task capsule network. Related Publication, Patent, and Funded Grant:
Rodney LaLonde, Drew Torigian, and Ulas Bagci. "Encoding Visual Attributes in Capsules for Explainable Medical Diagnoses." International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2020).
Rodney LaLonde and Ulas Bagci. "Capsules for Image Analysis." U.S. Patent Application 16/431,387; filed December 5, 2019.
Core X-Caps ideas were embedded into Dr. Ulas Bagci's NIH NCI R01 Grant. (Funded with high score for $2.1M; found “Extremely Innovative”).
Motivation for Explainable Diagnosis
Convolutional neural network (CNN) based systems have largely not adopted into clinical workflows. This hesitancy is seen in many high-risk applications (e.g., healthcare, military, security, transportation, finance, legal). The reluctance of adoption is cited as lack of trust, caused by the highly uninterpretable “black-box” nature of CNNs.
DARPA has invested billions over the past few years in “explainable” AI. Transportation Department wants XAI in self-driving cars. Patent & Trademark Office wants XAI for improving its patent approval process. Elham Tabassi, acting chief of staff at the National Institute of Standards and Technology (NIST) Information Technology Lab, and the lead author of the NIST report on a common set of AI principles, said this…
"We need them to be explainable, rather than just give an answer. They should be able to explain how they derive that prediction or to that decision… That goes a long way on increasing trust in the system."
Explainable vs. Interpretable
In the context of this work, interpretability deals with the post-hoc analysis of the inner workings of a model in relation to its predictions, such as Grad-CAM and saliency maps or blacking out parts of the input to see how it changes the output. Explainable methods on the other hand explicitly provide explanations for their predictions when making them. The argument for explainable predictions over post-hoc interpretations is this: Instead of a model predicting a picture is of a cat, and a researcher trying to break down the neural activation patterns of what parts of the image are activating which parts of the network, what end-users would really prefer, is for the model to explain its predictions just as a human would. We call this human-level explainability. When asking why is this a cat, a human would not vaguely point to regions of the image, or parts of their brain...

they would answer it’s a cat, because it has fur, and whiskers, and claws, etc. Humans explain their classifications of objects based on a taxonomy of object attributes, and if we want our models to be explainable at the human level, they should provide end-users with these same kinds of explanations.

This brings us to the research question for this chapter...
Can we build a capsule network to model specific visually-interpretable object attributes & form predictions based solely on their combination?
Explainable Lung Cancer Diagnosis
As an application of this research we chose lung cancer diagnosis. Lung cancer is a perfect application within medical imaging diagnosis, because radiologists already explain their predictions for nodule malignancy, based on a taxonomy of attributes, including subtlety, sphericity, margin, lobulation, spiculation, and texture.

An Explainable Multi-Task Capsule Network (X-Caps)
To solve this problem, we propose an explainable multi-task capsule network.

An object, in this case a lung nodule, is input to our three-layer 2D capsule network to form attribute prediction capsule vectors. Each of these vectors is supervised to encode a specific visually-interpretable attribute of the target object, where the dimensions of each vector capture the possible variations of that attribute over the dataset, and the magnitude of the vector represents the attribute presence, or in our case its score. Then, we predict the nodule’s malignancy by passing these visually-interpretable capsules through a linear function and apply a softmax activation to create a probability distribution over malignancy scores, while also passing them to a reconstruction branch to perform regularization. For creating these attribute capsules, unlike in CapsNet where parent capsules were mutually-exclusive (for example if class prediction is the digit 5 it cannot also be a 3), our parent capsules are not mutually-exclusive of each other, where a nodule can score high or low in each of the attribute categories. For this reason, we modify the dynamic routing algorithm to independently route information from children to parents through a “routing sigmoid” function:

Where the original “routing softmax” employed by CapsNet enforces a one-hot mapping of information from each child to parents, our proposed routing sigmoid learns a non-mutually-exclusive relationship between children and parents to allow multiple children to be emphasized for each parent, while the rest of the dynamic routing procedure following the same as in CapsNet.
Building-in Confidence Estimation
Typically, in lung nodule classification datasets, a minimum of three radiologists provide their scores on a scale of one to five for nodule malignancy. Previous studies in this area follow a strategy of averaging radiologists’ scores and then attempt to either regress this average or perform binary classification as above or below three. However, such approaches throw away valuable information about the agreement or disagreement amongst experts. To better model the uncertainty inherently present in the labels due to inter-observer variation, we propose to directly predict the distribution of radiologists’ scores by fitting a Gaussian function to the mean and variance as the ground-truth for our classification vector. This allows us to model the uncertainty present in radiologists’ labels and provide a meaningful confidence metric at test time to radiologists. Nodules with strong inter-observer agreement will produce a sharp peak as the ground-truth during training, in which case predictions with large variance (i.e. low confidence) will be punished. Likewise, for nodules with poor inter-observer agreement, we expect our network to output a more spread distribution and will be punished for strongly predicting a single class label, even if correct. At test, the variance in the predicted distribution provides radiologists with an estimate of the model’s confidence in that prediction.
Multi-Task Loss Formulation
X-Caps, being a multi-task framework, has 3 losses in its overall objective function. First, for the reconstruction branch, we choose to reconstruct only the nodule region of the input, masked by the ground-truth segmentation, then we compute the mean-squared error between this and the reconstruction branch output.

Next, for our 6 attribute predictions, we compute the mean-squared error between the network predictions and the normalized mean of radiologist scores for each attribute.

Lastly, for predicting malignancy, we compute the KL Divergence between a Gaussian distribution fit to the mean and variance of radiologist scores, and the softmax over our malignancy output prediction vector.

The total loss is the sum of these three loss functions.
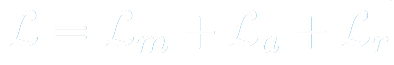
For simplicity we choose to set the loss balancing coefficients to 1 for all terms except the reconstruction branch, which was set to 0.5 to prevent over-regularizing the network. It’s worth noting briefly that engineering efforts spent to carefully tune these parameters, could lead to superior performance.
Experiments and Results
We performed experiments on the LIDC dataset, where at least 3 radiologists annotated 646 benign and 503 malignant nodules, excluding nodules of mean malignancy score exactly 3. Our method was compared against the state-of-the-art explainable CNN for lung cancer diagnosis, called HSCNN, which is a deep, dense, dual-path, 3D CNN, as well as two non-explainable 3D CNNs and the original CapsNet. The results of our experiments show, that supervising the attributes learned within the vectors of our capsule network significantly improved our performance over CapsNet, while a CNN-based method which built an identical explainable hierarchy of first predicting attributes then malignancy suffered from degraded performance compared to its non-explainable counterparts, as shown in the symbolic plot.

Here are the quantitative results of our experiments, where our simple 2D, 3-layer, X-Caps significantly outperformed the explainable HSCNN on predicting attribute scores, while also achieving higher malignancy prediction accuracy, with performance comparable to that of the non-explainable, deep, multi-crop or multi-scale 3D CNNs.

Conclusions and Discussion
Available studies for explaining DL models, typically focus on post hoc interpretations of trained networks, rather than attempting to build-in explainability. This is the first study for directly learning an interpretable feature space by encoding high-level visual attributes within the vectors of a capsule network to perform explainable image-based diagnosis. We approximate visually-interpretable attributes through individual capsule types, then predict malignancy scores directly based only on these high-level attribute capsule vectors, in order to provide malignancy predictions with explanations at the human-level, in the same language used by radiologists. Our proposed multi-task explainable capsule network, X-Caps, successfully approximated visual attribute scores better than the previous state-of-the-art explainable diagnosis system, while also achieving higher diagnostic accuracy. We hope our work can provide radiologists with malignancy predictions which are explained via the same high-level visual attributes they currently use, while also providing a meaningful confidence metric to advise when the results can be more trusted, thus allowing radiologists to quickly interpret and verify our predictions. Lastly, we believe our approach should be applicable to any image-based classification task where high-level attribute information is available to provide explanations about the final prediction.
Chapter 5: Dissertation Conclusion & Future Work

Final Conclusions
In Chapter 2, we introduced the first ever capsule-based segmentation network in the literature, SegCaps, while producing several novel advancements, including a locally-constrained dynamic routing algorithm, transformation matrix sharing, “deconvolutional” capsules, extension of the reconstruction regularization to segmentation, and a deepencoder-decoder capsule architecture. We validated the effectiveness and efficiency of SegCaps in the largest ever study in pathological lung segmentation, and the only showing results on pre-clinical subjects utilizing deep learning methods, and showed that SegCaps consistently outperforms all state-of-the-art CNN-based approaches while only using a small fraction of the total parameters of these much larger networks. Further, our additional experiments conducted on fluorescein angiogram videos, and rotations and reflections of natural images gives compelling evidence for the advantages of a capsule-based segmentation method over CNN-based methodologies.
In Chapter 3, we introduced a deep capsule network for medical image diagnosis, D-Caps,which was able operate on high-dimensional imaging data thanks to our novel capsule-average pooling algorithm and showed significantly improved results over the state-of-the-art CNN-based method on the limited training data and high intra-class variation present in the Mayo Polyp dataset, where we diagnosed non-localized colorectal polyps from single images.
Lastly, in Chapter 4, we created a novel multi-task explainable capsule network, X-Caps,which learned to encode visual attributes within its vectors, to provide malignancy predictions with the same high-level explanations used by human-expert radiologists. X-Caps utilizesa novel routing sigmoid function to independently route information from child capsules to their non-mutually-exclusive parents, while being trained directly on the distribution of expert labels to model inter-observer agreement and provide a meaningful metric of model over- or under-confidence supervised by human-experts’ agreement. We demonstrate a simple 2D, 3-layer, capsule network can outperform a state-of-the-art deep, dense, dual-path, 3D CNN at capturing visually-interpretable high-level attributes and malignancy prediction, while providing malignancy prediction scores comparable to non-explainable 3D CNNs.
Future Directions
We hope this dissertation will inspire future research into capsule networks, and to encourage this, we note a few potential research directions.
From the technical side:
Hinton cited the dynamic routing algorithm as critical to finally make capsule networks a reality; however, there is strong evidence that the current iterative-based routing mechanisms in the literature are significantly sub-optimal, and we would encourage future investigation in this area.
Further, we would like to encourage future researchers to consider capsule networks within the domain of representation learning and disentanglement where there are some interesting parallels, specifically to concept-vector methods in particular.
From the application side:
We would like to draw attention to the fact that no studies have been able to show a capsule network achieving comparable results to CNNs on large-scale classification tasks such as ImageNet or on object detection tasks such as MS COCO.
In our personal future work, we recently submitted a study which introduced a capsule network able to perform object detection on MS COCO, which utilizes new deformable capsules, a unique split-capsule detection head, and a novel squeeze-and-excitation inspired non-iterative dynamic routing algorithm.
Acknowledgements
A huge thank you to my adviser Dr. Bagci for all the advice, guidance, and support over the last several years.
Thank you to Dr. Shah for co-advising me during my first year and the great experience he instilled into me.
To the other members of my committee, Dr. Mahalanobis and Dr. Wallace, thank you for your commitment and support.
NIH Funding Subcontract (Dr. Wallace, Dr. Bagci), FDOH Funding, ORC Fellowship.
To all my lab mates, thanks for keeping me sane and all the great discussions on research ideas.
To my parents and brother, for their unending support and love.
To my fiance, for her love and support through all the sleepless nights of working late in the lab.


Dear Dr. LaLonde
In Segcaps network code, according to version 1.13.1 for tensorflow, it is not applicable in many GPU systems, is there a solution? Thankful
Excellent presentation, you have given a really intuitive description of the work and made this easier to understand - thank you!
I am undertaking a research masters degree and will be looking in depth at capsule networks and chest X ray applications so will be reading all your work in detail.